Research
Research Thrust I: Incorporating Biological Priors for Better Data Representations
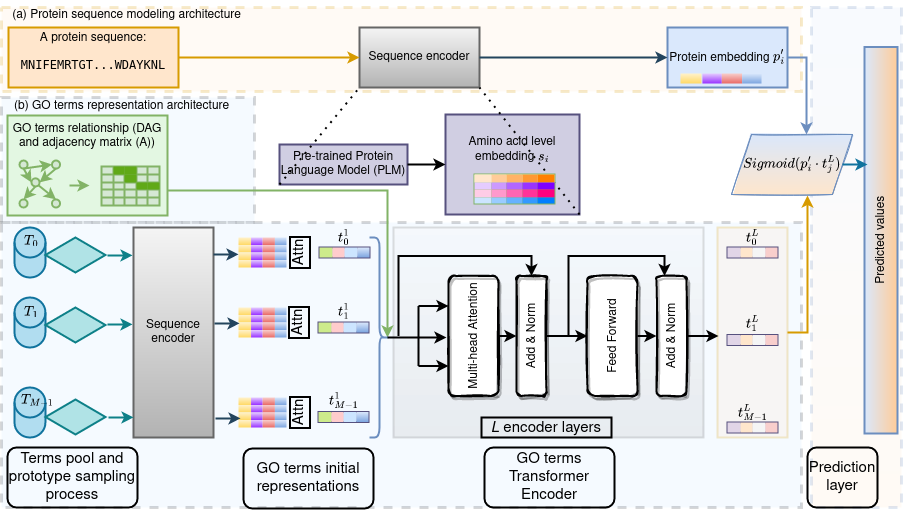
Large molecular language models are emerging as a powerful tool to learn and extract useful information from molecular sentences and linking them to functions. However, these models, typically trained on sequences alone, miss opportunities to integrate data from heterogeneous sources. Inspired by molecular evolution and the relationship between structure, function and properties, we propose models that incorporate multiple molecular representations. As an example, we incorporate mutual functional terms’ relationships as biological priors to annotate proteins from only sequence information. We attribute two key contributions: a novel method that utilizes the transformer architecture in two ways, one for encoding protein sequence and another for encoding functional hierarchy, and a deep investigation of different ways of constructing training and testing datasets. The learned sequence and functional terms’ representations are combined and utilized for multi-label Gene Ontology (GO) term classification. The method is shown superior over three recent representative GO prediction methods. The work also reveals that existing approaches under- or over-estimate the generalizability of a model. A novel approach is proposed to address these issues, resulting in a new benchmark dataset to rigorously evaluate, compare and advance the methods. This work has appeared in Biomolecules, 2022 (impact factor 5.80).
Research Thrust II: Integrating Physical Processes beyond Biological Priors
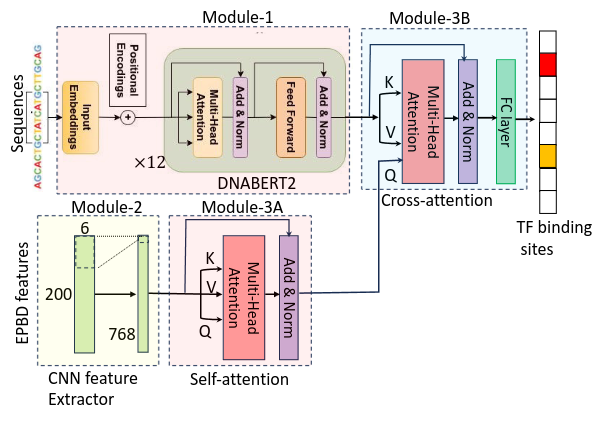
Decades of simulation-derived physical analyses in biology have been somewhat overlooked with the rise of deep learning. We posit that certain knowledge might not be fully captured by DL models alone, necessitating physical guidance. To bridge this gap, we propose to incorporate physics-based priors into biological foundation models, as such we apply physics-based DNA features to guide a genomic foundation model for transcription factor (TF) binding prediction tasks. We developed a multi-modal DL model, EPBDxDNABERT-2, to ascertain the precise relationship between TF-DNA binding. The key contribution of this research is the integration of physical processes and guidance to the foundation model through the cross attention (Fig. 3). Trained on the experimental data encompassing 161 distinct TFs and 91 human cell types EPBDxDNABERT-2 significantly improves the prediction of over 660 TF-DNA, with an increase in the AUROC metric of up to 9.6% when compared to the baseline model that does not leverage DNA biophysical properties. Extended analysis on an in vitro dataset of 215 TFs from 27 families demonstrates the effectiveness of the DNA breathing features with established frameworks. This work has appeared in the Nucleic Acid Research, 2024 (impact factor 16.6).
Research Thrust III: Exposing Fundamental Properties and Organization of the Representation Space
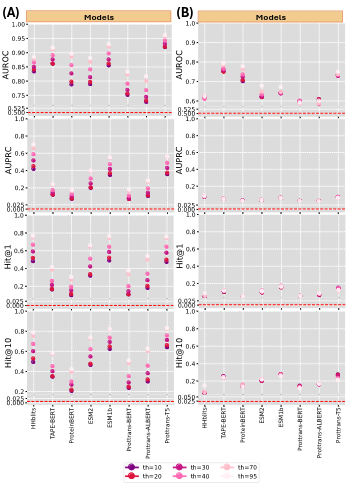
Over the years, numerous computational methods have been developed to accurately model the language of life. To enhance explainability in language models, we investigated the properties of the learned embedding space for protein molecular domains, concentrating on two challenging areas: remote homology prediction and mutation effect analysis. We address this by evaluating PLMs on remote homology prediction, where identifying remote homologs from sequence information alone requires structural knowledge, especially in the “twilight zone” of very low sequence identity. Through rigorous testing, we profile the performance of PLMs ranging from millions to billions of parameters in a zero-shot setting. Our findings indicate that PLMs still struggle in the twilight zone, that is, the current PLMs have not sufficiently learned protein structure to address remote homology prediction when sequence signals are weak. This has appeared in Bioinformatics Advances, 2024 (impact factor 4.4).